Research Centre for Computational Health
About us
The Research Centre for Computational Health addresses problems in medicine and biology using data-driven and mechanistic modeling. Important tools are machine learning for image and signal analysis, graphical networks, parameter estimation for differential equation systems and physiological simulation.
Biomedical Simulation
The research group specializes in modeling biological and medical systems. New approaches are developed to simulate physiological processes and to predict pathological changes. In particular, in-depth knowledge of biological/physiological processes is incorporated into multi-physics simulations.
The group develops algorithms for parameter and uncertainty estimation of physically motivated stochastic models. In particular, machine learning methods are combined with Bayesian modeling for dimensionality reduction. These methods are widely used in medicine and life sciences.
Group leader: Prof. Dr. Sven Hirsch | Learn more about the research group Biomedical Simulation
Medical Image Analysis
The research group applies machine learning techniques to interpret medical image data. This way, features are extracted for the characterization of disease patterns and for use as diagnostic markers. Of particular interest are the radiomic and morphological analysis of diagnostic medical imaging data. The group pursues the goal of establishing reproducible, image-based biomarkers by means of explainable artificial intelligence and ensuring their clinical utility.
Medical Data Modelling
The research group uses statistical and machine learning methods to model and uncover causal relations in medical data, especially to study pathophysiological processes. Categorial patient data and imaging date, e.g. magnetic resonance imaging, are processed to extract clinical knowledge.
Biosignal Analysis & Digital Health
The research group studies data from wearables and biosensors using time series analysis and combines them with biological-physical models to robustly characterize physiological systems. These data sources are used for Patient Reported Outcomes in clinical practice and for the further development of patient-centered medicine.
Group leader: Dr. Samuel Wehrli | Learn more about the research group Biosignal Analysis & Digital Health
Teaching Activities
The courses offered by the centre of Computational Health cover topics in mathematical and physical modeling. At Bachelor level it is responsible for the specialization Digital Health. The specialization teaches physics and mathematics for other BSc programs at the department. In the MSc Applied Computational Life Sciences and the PhD program Data Science advanced courses in modeling of complex systems are taught.
Projects
-
Digital Health Zurich – A practice lab for patient-centred clinical innovation
Digital Health Zurich researches digital health solutions in the hospital context and implements them efficiently and with practical relevance. Core topics are Patient Reported Outcome Measure (PROMs), remote monitoring, integrated care and related technologies as well as empowerment of patients and staff. Our ...
-
Modeling of multicentric and dynamic stroke health data
The aim of the project is the further development of probabilistic and dynamic modeling of diseases in the form of Bayesian networks in the field of digital health as a central strategic pillar of the newly formed specialist group "Medical Image Analysis and Data Modeling" of the "Computational Health" focus. To ...
-
Feature Learning for Bayesian Inference
The goal of this project is to use interpretable Machine Learning (ML) to find low-dimensional features in high-dimensional noisy data generated by (i) stochastic models or (ii) real systems. In both cases, the problem is to disentangle the effect of high-dimensional disturbances, such as noise or unobserved inputs, ...
-
OSR4H – Open Set Recognition for Hematology
Development of a Proof of Concept for visual Open Set Recognition (OSR) algorithms applied to a Hematology task, the classification of white blood cells.
-
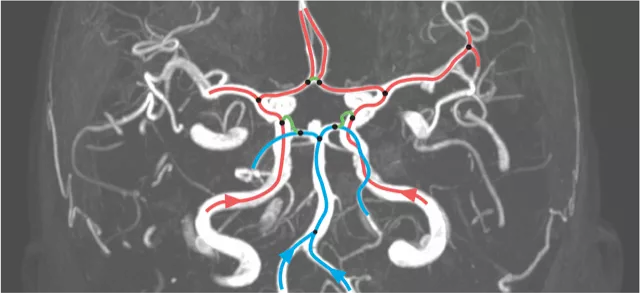
Deep Brain Vessel Profiler
The architecture of the supplying brain blood vessels is believed to impact the occurrence and severity of common cerebrovascular diseases such as ischemic strokes or intracranial aneurysms. In this project, we study methods to efficiently quantify the variability in human cerebral vasculature and how this ...
Publications
-
Morel, Sandrine; Hostettler, Isabel C.; Spinner, Georg R.; Bourcier, Romain; Pera, Joanna; Meling, Torstein; Alg, Varinder; Houlden, Henry; Bakker, Mark; van’t Hof, Femke; Rinkel, Gabriel; Foroud, Tatiana; Lai, Dongbing; Moomaw, Charles; Worrall, Bradford; Caroff, Jildaz; Constant-dits-Beaufils, Pacôme; Karakachoff, Matilde; Rimbert, Antoine; Rouchaud, Aymeric; Gaal-Paavola, Emilia; Kaukovalta, Hanna; Kivisaari, Riku; Laakso, Aki; Jahromi, Behnam; Tulamo, Riikka; Friedrich, Christoph; Dauvillier, Jerome; Hirsch, Sven; Isidor, Nathalie; Kulcsàr, Zolt; Lövblad, Karl; Martin, Olivier; Machi, Paolo; Mendes Pereira, Vitor; Rüfenacht, Daniel; Schaller, Karl; Schilling, Sabine; Slowik, Agnieszka; Jaaskelainen, Juha; von und zu Fraunberg, Mikael; Jiménez-Conde, Jordi; Cuadrado-Godia, Elisa; Soriano-Tárraga, Carolina; Millwood, Iona; Walters, Robin; The @neurIST project; The ICAN Study Group; Genetics and Observational Subarachnoid Haemorrhage (GOSH) Study Investigators; International Stroke Genetics Consortium (ISGC); Kim, Helen; Redon, Richard; Ko, Nerissa; Rouleau, Guy; Lindgren, Antti; Niemelä, Mika; Desal, Hubert; Woo, Daniel; Broderick, Joseph; Werring, David; Ruigrok, Ynte; Bijlenga, Philippe,
2022.
Intracranial aneurysm classifier using phenotypic factors : an international pooled analysis.
Journal of Personalized Medicine.
12(9), pp. 1410.
Available from: https://doi.org/10.3390/jpm12091410
-
Albert, Carlo; Ulzega, Simone; Ozdemir, Firat; Perez-Cruz, Fernando; Mira, Antonietta,
2022.
Learning summary statistics for Bayesian inference with autoencoders.
SciPost Physics Core.
5(3), pp. 043.
Available from: https://doi.org/10.21468/SciPostPhysCore.5.3.043
-
Juchler, Norman; Bijlenga, Philippe; Hirsch, Sven,
2022.
Modeling the location-dependency of aneurysm shape : a morphometric comparative study [paper].
In:
Nithiarasu, Perumal; Vergara, Christian, eds.,
CMBE 2022 : 7th International Conference on Computational & Mathematical Biomedical Engineering.
7th International Conference on Computational and Mathematical Biomedical Engineering (CMBE22), Milan, Italy, 27-29 June 2022.
Computational & Mathematical Biomedical Engineering.
pp. 703-705.
Available from: https://doi.org/10.21256/zhaw-25388
-
Spinner, Georg R.; Delucchi, Matteo; Morel, Sandrine; Bijlenga, Philippe; Hirsch, Sven,
2022.
In:
Nithiarasu, Perumal; Vergara, Christian, eds.,
CMBE 2022 : 7th International Conference on Computational & Mathematical Biomedical Engineering.
7th International Conference on Computational and Mathematical Biomedical Engineering (CMBE22), Milan, Italy, 27-29 June 2022.
Computational and Mathematical Biomedical Engineering.
pp. 72-75.
-
Juchler, Norman; Bijlenga, Philippe; Hirsch, Sven,
2022.
The role of shape for aneurysm risk assessment [paper].
In:
Nithiarasu, Perumal; Vergara, Christian, eds.,
CMBE 2022 : 7th International Conference on Computational & Mathematical Biomedical Engineering.
7th International Conference on Computational and Mathematical Biomedical Engineering (CMBE22), Milan, Italy, 27-29 June 2022.
Computational & Mathematical Biomedical Engineering.
pp. 84-86.
Available from: https://doi.org/10.21256/zhaw-25386